Step 2: Run and verify the output of simaaiprocessmla MLA process
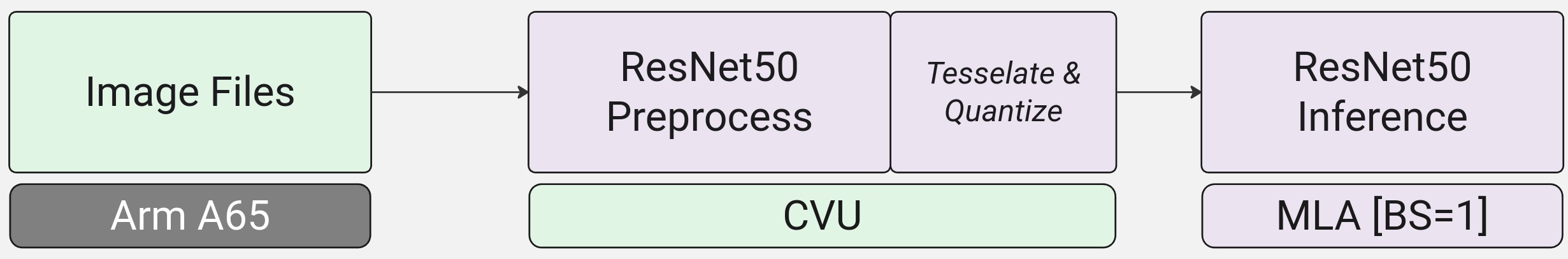
In this section we will explore running through the MLA plugin in order to run the ML model.
We have two options going forward:
We can only run the MLA plugin to ensure that we are getting the right output. If so, we will use the
simaaisrcwith the output/tmp/generic_preproc-001.outfrom the previous step as input and feed directly into thesimaaiprocessmlaplugin.We can simply expand the pipeline to now include the MLA step. In this guide, we will go with this step.
Before running the simaaiprocessmla plugin to perform inference on the MLA, we need to configure the json file for the plugin and ensure that we have saved the model locally on the board.
Copy the model to the MLSoC
Copy the quantized and compiled model from the Palette docker on the host machine to the MLSoC:
sima-user@docker-image-id$ scp models/compiled_resnet50/quantized_resnet50_mpk.tar.gz sima@<IP address of MLSoC>:/home/sima/resnet50_example_app/models/
From the MLSoC shell prompt, lets extract the contents:
davinci:~/resnet50_example_app/models$ tar xvf quantized_resnet50_mpk.tar.gz
quantized_resnet50_stage1_mla.lm
quantized_resnet50_mpk.json
quantized_resnet50_stage1_mla_stats.yaml
Creating the JSON configuration file
On the MLSoC, create the JSON configuration in /home/sima/resnet50_example_app/app_configs.
davinci:~/resnet50_example_app/app_configs$ ls
Run the following command:
echo '{
"version" : 0.1,
"node_name" : "mla-resnet",
"simaai__params" : {
"params" : 15,
"index" : 1,
"cpu" : 4,
"next_cpu" : 1,
"out_sz" : 1008,
"no_of_outbuf" : 1,
"batch_size" : 1,
"batch_sz_model" : 1,
"in_tensor_sz": 0,
"out_tensor_sz": 0,
"ibufname" : "generic_preproc",
"model_path" : "/home/sima/resnet50_example_app/models/quantized_resnet50_stage1_mla.lm",
"debug" : 0,
"dump_data" : 1
}
}' > simaaiprocessmla_cfg_params.json
The GStreamer string update
Let’s update the previous run_pipeline.sh script to include our new plugin.
#!/bin/bash
# Constants
APP_DIR=/home/sima/resnet50_example_app
DATA_DIR="${APP_DIR}/data"
SIMA_PLUGINS_DIR="${APP_DIR}/../gst-plugins"
SAMPLE_IMAGE_SRC="${DATA_DIR}/golden_retriever_207_rgb.bin"
CONFIGS_DIR="${APP_DIR}/app_configs"
PREPROC_CVU_CONFIG_BIN="${CONFIGS_DIR}/genpreproc_200_cvu_cfg_app"
PREPROC_CVU_CONFIG_JSON="${CONFIGS_DIR}/genpreproc_200_cvu_cfg_params.json"
INFERENCE_MLA_CONFIG_JSON="${CONFIGS_DIR}/simaaiprocessmla_cfg_params.json"
# Remove any existing temporary files before running
rm /tmp/generic_preproc*.out
# Run the configuration app for generic_preproc
$PREPROC_CVU_CONFIG_BIN $PREPROC_CVU_CONFIG_JSON
# Run the application
export LD_LIBRARY_PATH="${SIMA_PLUGINS_DIR}"
gst-launch-1.0 -v --gst-plugin-path="${SIMA_PLUGINS_DIR}" \
simaaisrc mem-target=1 node-name="my_image_src" location="${SAMPLE_IMAGE_SRC}" num-buffers=1 ! \
simaaiprocesscvu source-node-name="my_image_src" buffers-list="my_image_src" config="$PREPROC_CVU_CONFIG_JSON" name="generic_preproc" ! \
simaaiprocessmla config="${INFERENCE_MLA_CONFIG_JSON}" name="mla_inference" ! \
fakesink
To run the application:
davinci:~/resnet50_example_app$ sudo sh run_pipeline.sh
Password:
Completed SIMA_GENERIC_PREPROC graph configure
** Message: 04:37:40.073: Num of chunks 1
** Message: 04:37:40.073: Buffer_name: my_image_src, num_of_chunks:1
(gst-launch-1.0:2398): GLib-GObject-CRITICAL **: 04:37:40.084: g_pointer_type_register_static: assertion 'g_type_from_name (name) == 0' failed
(gst-launch-1.0:2398): GLib-GObject-CRITICAL **: 04:37:40.085: g_type_set_qdata: assertion 'node != NULL' failed
(gst-launch-1.0:2398): GLib-GObject-CRITICAL **: 04:37:40.085: g_pointer_type_register_static: assertion 'g_type_from_name (name) == 0' failed
(gst-launch-1.0:2398): GLib-GObject-CRITICAL **: 04:37:40.086: g_type_set_qdata: assertion 'node != NULL' failed
Setting pipeline to PAUSED ...
** Message: 04:37:40.093: Initialize dispatcher
** Message: 04:37:40.094: handle: 0xa3b295b0, 0xffffa3b295b0
** Message: 04:37:41.238: Loaded model from location /data/simaai/building_apps_palette/gstreamer/resnet50_example_app/models/quantized_resnet50_stage1_mla.lm, model:hdl: 0xaaaae079eaa0
** Message: 04:37:41.242: Filename memalloc = /data/simaai/building_apps_palette/gstreamer/resnet50_example_app/data/golden_retriever_207_rgb.bin
Pipeline is PREROLLING ...
Pipeline is PREROLLED ...
Setting pipeline to PLAYING ...
Redistribute latency...
New clock: GstSystemClock
Got EOS from element "pipeline0".
Execution ended after 0:00:00.001474163
Setting pipeline to NULL ...
Freeing pipeline ...
You will see the output of the CVU preprocess and the MLA inference in the /tmp/ folder:
davinci:~/resnet50_example_app$ ls /tmp/*.out
generic_preproc-001.out mla-resnet-1.out
Verifying the output
Just like the input of the MLA needs to be quantized and tesselated, the output of the MLA is still quantized and tesselated.
Thus, any reference we are going to compare against, also needs to be in the same form, or, we must dequantize and detesselate before verifying the output.
Let’s first take a look at the output from the plugin:
davinci:~/resnet50_example_app$ hexdump -C /tmp/mla-resnet-1.out
00000000 80 80 80 80 80 80 80 80 80 80 80 80 80 80 80 80 |................|
*
000000c0 80 80 80 80 80 80 80 80 80 80 80 80 80 80 80 7f |................|
000000d0 81 80 80 80 80 80 80 80 81 80 80 80 80 80 80 80 |................|
000000e0 80 80 80 80 80 80 80 80 80 80 80 80 80 80 80 80 |................|
*
000003e0 80 80 80 80 80 80 80 80 00 00 00 00 00 00 00 00 |................|
000003f0
As noted earlier, this output should be interpreted as 1008 values representing the output of the softmax function from the network of type int8 (2’s complement).
Note
Why 1008 values and not 1000 as expected for the ResNet50 output? This is just due to memory alignment requirements (tesselation) for the MLA.
When the output is detesselated, we will again have the output having 1000 values.
Note
The * between values of the hexdump signify that there are n lines before that contain repeated values.
In order to view the entire output, you can use the -v flag:
hexdump -C -v /tmp/mla-resnet-1.out
Using a Python on the MLSoC, let’s manually dequantize the output in order to see if the top 3 results match our expectations:
davinci:~/resnet50_example_app$ vi print_mla_top_output_indices.py
Copy the following inside the script:
import numpy as np
# Step 1: Read the binary file as int8 values
mla_data = np.fromfile('/tmp/mla-resnet-1.out', dtype=np.int8)[9:]
# Step 2: Dequantize (values from *_mpk.json for the output dequantization node)
dequantize_scale, dequantized_zero_point = 255.02200010497842, -128
dequantized_data = (mla_data - dequantized_zero_point).astype(np.float32) / dequantize_scale
# Step 3: Find the indices of the top 3 largest values
top_3_indices = np.argpartition(dequantized_data, -3)[-3:]
# Step 4: Sort the top 3 indices by the actual values (descending)
top_3_indices = top_3_indices[np.argsort(-dequantized_data[top_3_indices])]
# Print the results
print("Top 3 largest values and their indices:")
for idx in top_3_indices:
print(f"Index: {idx}")
Note
The values in the script can be extracted from the compiled .tar.gz *_mpk.json found under:
dequantize_scale =
plugins[5] → config_params → params → channel_params[0][0]dequantized_zero_point =
plugins[5] → config_params → params → channel_params[0][1]
mla_data is gathered without the first 8 pixels ([9:]) in order to remove 0’s that are a result of tesselation.
Run the script to take a look at the highest value classes to get an idea if the expected 207 class is the top class:
davinci:~/resnet50_example_app$ python3 print_mla_top_output_indices.py
Top 3 largest values and their indices:
Index: 207
Index: 199
Index: 332
Excellent, that is what we expected.
Conclusion and next steps
In this section, we:
Went through the steps of setting up the
simaaiprocessmlaplugin to run inference using a model that was compiled using the ModelSDK.Ran and verified the output given the
dumpof the plugin as configured in the JSON and the output from our python reference application
Next, we will add another CVU graph in order to detesselate and dequantize the output from the MLA.